In-Silico Molecular Docking and ADMET Prediction of Natural Compounds In Oncom
Main Article Content
Abstract
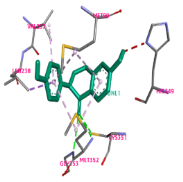
Oncom is a traditional fermented cuisine from West Java, Indonesia, produced through the process of mold fermentation. Oncom is categorized into black oncom (BO) and red oncom (RO) based on the colour of the spore of the molds used in their fermentation. Both types of oncom are commonly found in traditional markets in West Java, Indonesia. This study aimed to explore and predict the diverse activities of bioactive compounds from both oncom by analyzing their interactions with multiple enzymes using the in silico method. The ethanol extracts of oncom were analyzed using liquid chromatography-high-resolution mass spectrometry (LC-HRMS). The absorption, distribution, metabolism, excretion, and toxicity (ADMET) predictions of the substances were performed using pkCSM and ProTox-II network servers, along with Lipinski’s rule of five screening. Molecular docking was performed on β-lactamase (PDB ID: 1LLB), cytochrome P450 (PDB ID: 1JIP), human COX-2 (PDB ID: 3LN1), SARS-COV-2 protease (PDB ID: 6LU7), and protein kinase (PDB ID: 1MV5) using AutoDockTools. The LC-HRMS analysis revealed the presence of 64 compounds in oncom extracts. ADMET together with Lipinski’s guidelines screening, predicted that nine of the 64 compounds showed promising biological activities. Daidzein exhibited a higher Gibbs free energy (∆G) of -6.44 kcal/mol for 1LLB compared to clavulanic acid (-5.07 kcal/mol) and -5.91 kcal/mol for 1MV5 in contrast to verapamil (-4.04 kcal/mol). The findings indicate that daidzein exhibits potential inhibitory activity of bacterial infections and cancer cells.
Downloads
Article Details
Section

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
How to Cite
References
Niazi SK and Mariam Z. Computer-Aided Drug Design and Drug Discovery: A prospective Analysis. Pharmaceuticals. 2024; 17(1):1-22. doi: 10.3390/ph17010022
Dhudum R, Ganeshpurkar A, Pawar A. Revolutionizing Drug Discovery: A Comprehensive Review of AI Applications. Drug Drug Candidates. 2024; 3(1):148-171. doi: 10.3390/ddc3010009
Stielow M, Witczyńska A, Kubryń N, Fijałkowski Ł, Nowaczyk J, Nowaczyk A. The Bioavailability of Drugs—The Current State of Knowledge. Molecules. 2023; 28(8038):1-19. doi: 10.3390/molecules28248038
Rai M, Singh AV, Paudel N, Kanase A, Falletta E, Kerkar P, Heyda J, Barghash RF, Singh SP, Soos M. Herbal Concoction Unveiled: A Computational Analysis of Phytochemicals’ Pharmacokinetic and Toxicological Profiles Using Novel Approach Methodologies (NAMs). Curr Res Toxicol. 2023; 5100118):1-13. doi: 10.1016/j.crtox.2023.100118
Nhlapho S, Nyathi MHL, Ngwenya BL, Dube T, Telukdarie A, Munien I, Vermeulen A, Chude-Okonkwo UAK. Druggability of Pharmaceutical Compounds Using Lipinski Rules with Machine Learning. Pharm. Sci. 2024; 3(4):177-192. doi: 10.58920/sciphar0304264
Bitew M, Desalegn T, Demissie TB, Belayneh A, Endale M, Eswaramoorthy R. Pharmacokinetics and Drug-likeness of Antidiabetic Flavonoids: Molecular Docking and DFT Study. PLoS One. 2021; 16(12):1-22. doi: 10.1371/journal.pone.0260853
Goodsell DS, Zardecki C, Di Costanzo L, Duarte JM, Hudson BP, Persikova I, Segura J, Shao C, Voight M, Westbrook JD, Young JY, Burley SK. RCSB Protein Data Bank: Enabling Biomedical Research and Drug Discovery. Protein Sci. 2020; 29(1):52-65. doi: 10.1002/pro.3730
Singh N, Vayer P, Tanwar S, Poyet J-L, Tsaioun K, Villoutreix BO. Drug Discovery and Development: Introduction to the General Public and Patient Groups. Front Drug Discov. 2023; 3:1-11. doi: 10.3389/fddsv.2023.1201419
Wardana AP, Aminah NS, Kristanti AN, Fahmi MZ, Abdjan MI, Sucipto TH. Antioxidant, Anti-inflammatory, Antiviral and Anticancer Potentials of Zingiberaceae Species Used as Herbal Medicine in Indonesia. Trop J Nat Prod Res. 2024; 8(9):8393-8399. doi: 10.26538/tjnpr/v8i9.22
Shah M, Patel M, Shah M, Patel M, Prajapati M. Computational Transformation in Drug Discovery: A Comprehensive Study on Molecular Docking and Quantitative Structure Activity Relationship (QSAR). Intell Pharm. 2024; 2(5):589-595. doi: 10.1016/j.ipha.2024.03.001
Tamang JP. Ethnic Fermented Foods and Alcoholic Beverages of Asia. 1st ed. New Delhi: Springer India. 2016; 409p. doi: 10.1007/978-81-322-2800-4
Andayani SN, Lioe HN, Wijaya CH, Ogawa M. Umami Fractions Obtained from Water-soluble Extracts of Red Oncom and Black Oncom—Indonesian Fermented Soybean and Peanut Products. J Food Sci. 2020; 85(3):657-665. doi: 10.1111/1750-3841.14942
Surya R, Romulo A. Antioxidant Profile of Red Oncom, an Indonesian Traditional Fermented Soyfood. Food Res. 2023; 7(4):204-210. doi: 10.26656/fr.2017.7(4).650
Wu F, Zhou Y, Li L, Shen X, Chen G, Wang X, Liang X, Tan M and Huang Z. Computational Approaches in Preclinical Studies on Drug Discovery and Development. Front Chem. 2020; 8(726):1-32. doi: 10.3389/fchem.2020.00726
OECD. Acute Oral Toxicity – Fixed Dose Procedure (chptr). OECD Guid Test Chem. OECD Publishing, Paris. 2002;1-14. doi: 10.1787/9789264070943-en
Ijoma IK, Okafor CE, Ajiwe VIE. Computational Studies of 5-methoxypsolaren as Potential Deoxyhemoglobin S Polymerization Inhibitor. Trop J Nat Prod Res. 2024; 8(10):8835-8841. doi: 10.26538/tjnpr/v8i10.28
Illian DN, Widiyana AP, Hasana AR, Maysarah H, Al Mustaniroh SS, Basyuni M. In Silico Approach: Prediction of ADMET, Molecular Docking, and QSPR of Secondary Metabolites in Mangroves. J Appl Pharm Sci. 2022; 12(11):021-029. doi: 10.7324/JAPS.2022.121103
Ernawati T, Artanti N, Kumiawan YD. In Silico and In Vitro Anti-cancer Activity against Breast Cancer Cell Line MCF-7 of Amide Cinnamate Derivatives. J Appl Pharm Sci. 2024; 14(3):102-107. doi: 10.7324/JAPS.2024.162177
Fadilah F, Andrajati R, Arsianti A, Paramita RI, Erlina L, Istiadi KA, Yanuar A. Synthesis and In Vitro Activity of Eugenyl Benzoate Derivatives as BCL-2 Inhibitor in Colorectal Cancer with QSAR and Molecular Docking Approach. Asian Pac J Cancer Prev. 2023; 24(9):2973-2981. doi: 10.31557/APJCP.2023.24.9.2973
Abdelli W, Hamed D. Molecular Docking and Pharmacokinetics Studies of Syzygium aromaticum Compounds as Potential SARS-CoV-2 Main Protease Inhibitors. Trop J Nat Prod Res. 2023; 7(11):5155-5163. doi: 10.26538/tjnpr/v7i11.18
Maharani DA, Adelina R, Aini AQ, Supandi. Molecular Docking and Dynamic Simulation of Erythrina fusca Lour Chemical Compounds Targeting VEGFR-2 Receptor for Anti-Liver Cancer Activity. J Kim Val. 2024; 10(1):106-114. doi: 10.15408/jkv.v10i1.35295
Zuhri UM, Purwaningsih EH, Fadilah F, Yuliana ND. Network Pharmacology Integrated Molecular Dynamics Reveals the Bioactive Compounds and Potential Targets of Tinospora crispa Linn. as Insulin Sensitizer. PLoS One. 2022; 17(6):1-15. doi: 10.1371/journal.pone.0251837
Kondža M, Brizić I, Jokić S. Flavonoids as CYP3A4 Inhibitors in Vitro. Biomedicines. 2024; 12(3):1-23. doi: 10.3390/biomedicines12030644
Omar AM, Aljahdali AS, Safo MK, Mohamed GA, Ibrahim SRM. Docking and Molecular Dynamic Investigations of Phenylspirodrimanes as Cannabinoid Receptor-2 Agonists. Molecules. 2023; 28(1):1-17. doi: 10.3390/molecules28010044
Sutherland JB, Bridges BM, Heinze TM, Adams MR, Delio PJ, Hotchkiss C, Rafii, F. Comparison of the Effects of Antimicrobial Agents from Three Different Classes on Metabolism of Isoflavonoids by Colonic Microflora Using Etest Strips. Curr Microbiol. 2012; 64(1):60-65. doi: 10.1007/s00284-011-0020-4
Buchmann D, Schultze N, Borchardt J, Böttcher I, Schaufler K, Guenther S. Synergistic Antimicrobial Activities of Epigallocatechin Gallate, Myricetin, Daidzein, Gallic Acid, Epicatechin, 3-Hydroxy-6-Methoxyflavone, and Genistein Combined with Antibiotics Against ESKAPE Pathogens. J Appl Microbiol. 2022; 132(2):949-963. doi: 10.1111/jam.15253.
Jagatap VR, Ahmad I, Sriram D, Kumari J, Adu DK, Ike BW, Ghai M, Ansari SA, Ansari IA, Wetchoua POM, Karpoormath R, Patel H. Isoflavonoid and Furanochromone Natural Products as Potential DNA Gyrase Inhibitors: Computational, Spectral, and Antimycobacterial Studies. ACS Omega. 2023; 8(18):16228-16240. doi: 10.1021/acsomega.3c00684.
Yang J, Xiao P, Sun J, Guo L. Anticancer Effects of Kaempferol in A375 Human Malignant Melanoma Cells are Mediated via Induction of Apoptosis, Cell Cycle Arrest, Inhibition of Cell Migration and Downregulation of m-TOR/PI3K/AKT Pathway. J BUON. 2018; 23(1):218-223. https://www.jbuon.com/archive/23-1-218.pdf
Kumar V and Chauhan SS. Daidzein Induces Intrinsic Pathway of Apoptosis along with ER α/β Ratio Alteration and ROS Production. Asian Pac J Cancer Prev. 2021; 22(2):603-610. doi: 10.31557/APJCP.2021.22.2.603
Márquez-Flores YK, Martínez-Galero E, Correa-Basurto J, Sixto-López Y, Villegas I, Rosillo MÁ, Cárdeno A, Alarcón-de-la-Lastra C. Daidzein and Equol: Ex Vivo and In Silico Approaches Targeting COX-2, iNOS, and the Canonical Inflammasome Signaling Pathway. Pharmaceuticals. 2024; 17(647):1-19. doi: 10.3390/ph17050647
Jia J, He R, Yao Z, Su J, Deng S, Chen K, Yu B. Daidzein Alleviates Osteoporosis by Promoting Osteogenesis and Angiogenesis Coupling. PeerJ. 2023; 11:(e16121):1-20. doi: 10.7717/peerj.16121.